In 2015, for the first time in my 30-year scientific career, I had a paper rejected by four journals without even being sent to review. Of course, I am used to journals reviewing then deciding it is not for them – that does happen, but not to even send to experts?! I was very surprised by this, particularly since I had been speaking about the work described in the paper for almost two years at UK and international meetings and the talks had always drawn a large and interested audience. The paper being thrown out by editors without consulting experts in the field was the second in our series about a 48-replicate RNA-seq experiment which I have described earlier in this blog and was the result of a collaboration with fantastic experimentalists (Mark Blaxter at Edinburgh, and Tom Owen-Hughes and Gordon Simpson at Dundee). Thankfully, the fifth journal, RNA, sent it to review, and it was published on 16th May 2016. (Schurch, et al, 2016, RNA).
It was great to get the paper published in a good journal and especially nice to see that it stayed at No. 1 or No. 2 in the top accessed papers on the RNA website for six months (Figure 1 shows a snapshot – I did check this every week!).

This week (1st March 2017), I was doing some citation analysis and noticed to my surprise and pleasure that the RNA paper was rated by ISI Web of Science as “Hot” (Figure 2, while Google Scholar says it has 28 citations – I like Google Scholar!! ).
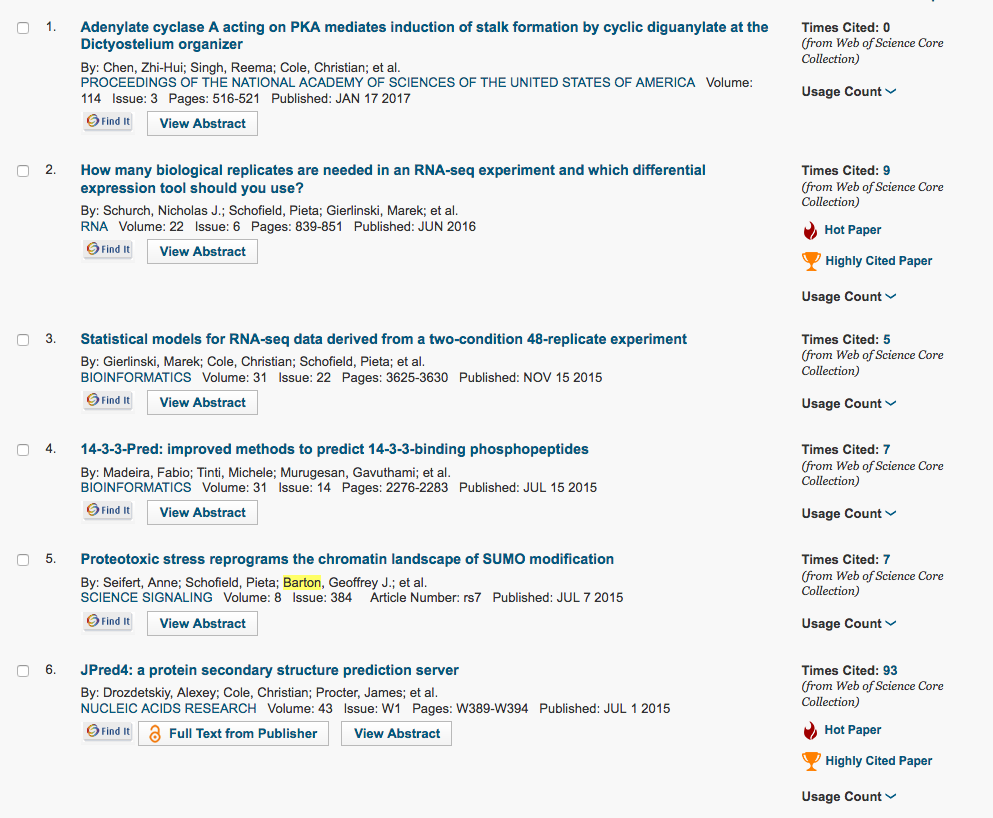
This means it is in the top 0.1% cited papers in the field of biochemistry or molecular biology for the last two years! In fact, there are currently only 28 “Hot” papers in this field from the whole UK. The work also generated the most interest on social media of any of the papers I have had a significant role in, with a current Altmetric score of 177 (Figure 3).
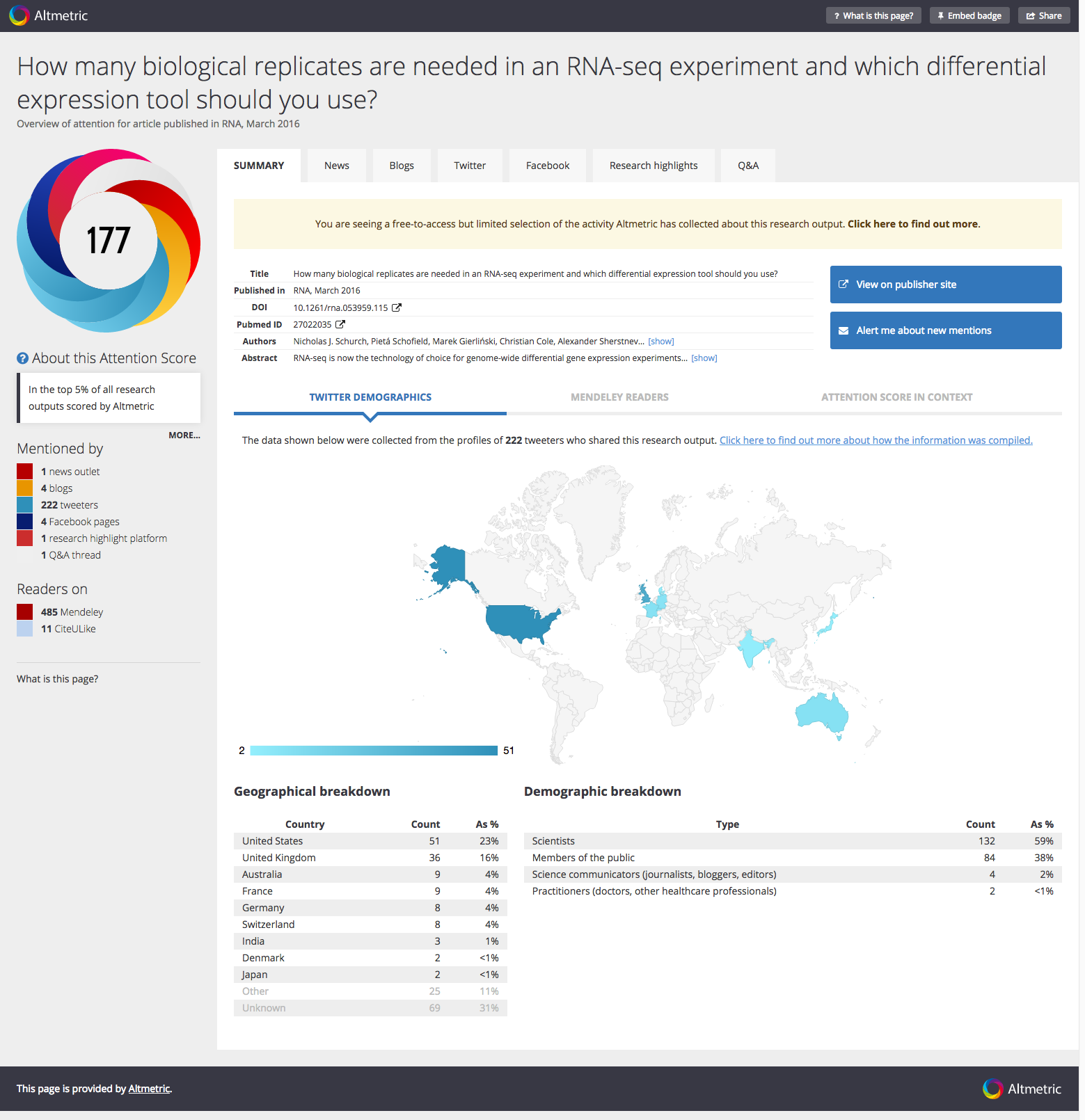
So, by any measure, this is a paper that was timely and continues to have impact!
The question then is: “Why did four journals consider it too boring to review?”.
I am not sure why eLife, Genome Biology, Genome Research and NAR would not review it, but will speculate a bit. I won’t quote the editor’s form rejections, but they all had a similar flavour that we have seen on other occasions: “Not of general interest.”, “Too technical.” “Appropriate for a more specialised journal…” To be fair to eLife, the original title and abstract for the paper were perhaps less “biologist friendly” than they could have been, but we fixed that before Genome Biology, Genome Research and NAR. To be fair to NAR, the editor was interested but did not think the paper fitted any of their categories of paper.
None of the editors said this explicitly but I did wonder if: “Not of general interest” was code for “not in human…”. Perhaps they thought our findings might not be relevant to a Eukaryote with a complex transcriptome? We also worried a bit about this, but our recent paper in Arabidopsis (Froussios et al, 2017; OK, not human, but certainly a Eukaryote with a complex transcriptome!) shows essentially the same result, albeit on fewer replicates. Another factor may have been due to us publishing the manuscript on the arXiv preprint server at the point of submission in 2015. Although all the journals say that they are happy with preprint submission, I wonder how happy some editors are with this? Of course, it may just be that they say this to everyone and so only respond to the authors who complain the loudest? I hope that is not the case.
Although it was annoying and time consuming at the time to have the paper rejected four times without review, I’m obviously very happy that the big, cross-institutional team that did this work was right about its importance! Lots of people from diverse fields have made positive comments to members of the team about how useful the paper is in their experimental design decisions. It is certainly good to feel that some of your work is appreciated!
I do wonder though if always aiming to publish in “good” journals is the right thing to do? Surely, it is more important that our work is visible and can be used by other scientists as early as possible? I will explore this some more in Part 2 of this blog, later…
0 Comments
1 Pingback